Looking for some help figuring out the rotation for conversion to instanced meshes.
The official three.js example can be seen here and code of my PDB Viewer can be seen here.
So far, I have managed to instance atoms, since they did not require rotation, but am stuck on converting bonds to instanced meshes and figuring out their rotation.
If anybody does have some suggestions then feel free to share it.
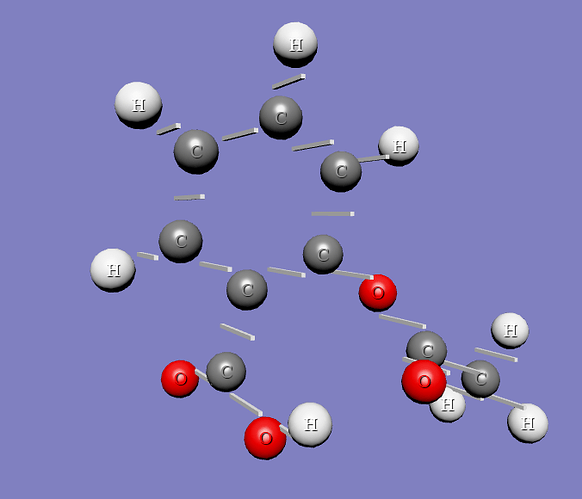
The pictures below reflect before and after situations.
Here is the part of the function which I am trying to convert to instanced mesh:
positions = geometryBonds.getAttribute( 'position' );
const start = new THREE.Vector3();
const end = new THREE.Vector3();
for ( let i = 0; i < positions.count; i += 2 ) {
start.x = positions.getX( i );
start.y = positions.getY( i );
start.z = positions.getZ( i );
end.x = positions.getX( i + 1 );
end.y = positions.getY( i + 1 );
end.z = positions.getZ( i + 1 );
start.multiplyScalar( 75 );
end.multiplyScalar( 75 );
const object = new THREE.Mesh( boxGeometry, new THREE.MeshPhongMaterial( { color: 0xffffff } ) );
object.position.copy( start );
object.position.lerp( end, 0.5 );
object.scale.set( 5, 5, start.distanceTo( end ) );
object.lookAt( end );
root.add( object );
}
and here is my attempt, which still does not have the rotation but produces the output as in the following picture:
positions = geometryBonds.getAttribute( 'position' );
const start = new THREE.Vector3();
const end = new THREE.Vector3();
const object = new THREE.InstancedMesh( boxGeometry, new THREE.MeshPhongMaterial( { color: 0xffffff } ), parseInt( positions.count / 2.0 ) );
for ( let i = 0; i < positions.count; i += 2 ) {
start.x = positions.getX( i );
start.y = positions.getY( i );
start.z = positions.getZ( i );
end.x = positions.getX( i + 1 );
end.y = positions.getY( i + 1 );
end.z = positions.getZ( i + 1 );
start.multiplyScalar( 75 );
end.multiplyScalar( 75 );
let m4 = new THREE.Matrix4();
m4.makeScale( 5, 5, start.distanceTo( end ) );
m4.setPosition( start.lerp( end, 0.5 ) );
object.setMatrixAt( parseInt( i / 2.0 ), m4 );
object.instanceMatrix.needsUpdate = true;
}
root.add( object );
All this so the number of draw calls could be reduced and some larger molecule examples could be easier loaded and manipulated, like examples from the RCSB Protein Data Bank(H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne, The Protein Data Bank (2000) Nucleic Acids Research 28: 235-242 Protein Data Bank | Nucleic Acids Research | Oxford Academic).